Researchers visualize how proteins self-assemble
June 11, 2012
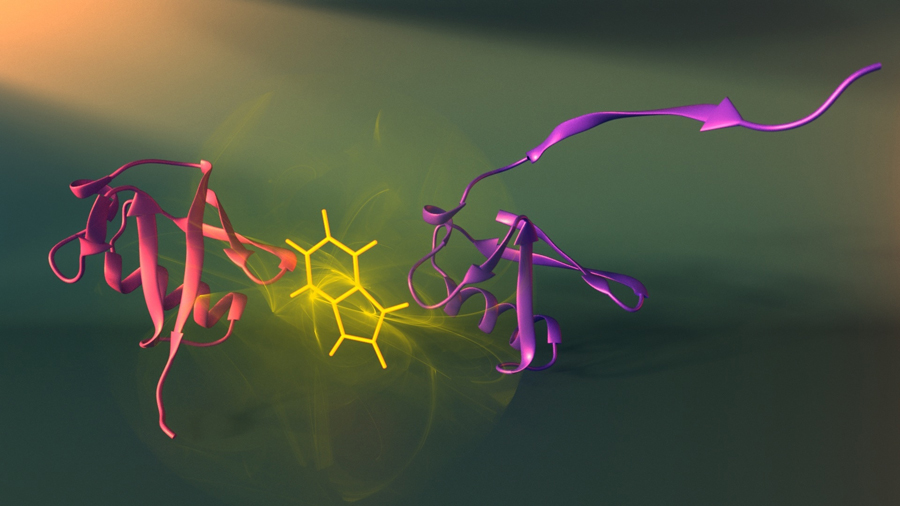
Two assembly stages (purple and red) of the protein ubiquitin and the fluorescent tryptophan probe (yellow) used to visualize these stages (credit: Peter Allen/University of Montreal/Nature Structural and Molecular Biology)
University of Montreal researchers have developed a new approach to visualizing how proteins assemble that may aid our understanding of diseases such as Alzheimer’s and Parkinson’s, which are caused by errors in assembly.
It could also help bioengineers design new molecular machines.
“To understand how a protein goes from a linear chain to a unique assembled structure, we need to capture snapshots of its shape at each stage of assembly,” said Dr. Alexis Vallée-Bélisle, first author of the study. “The problem is that each step exists for a fleetingly short time and no available technique enables us to obtain precise structural information on these states within such a small time frame.
The researchers integrated fluorescent probes throughout the linear protein chain to detect the structure of each stage of protein assembly, step by step.
“Understanding how a protein goes from being one thing to becoming another is the first step towards understanding and designing protein nanomachines for biotechnologies such as medical and environmental diagnostic sensors,” Vallée-Bélisle said.
This research was supported by the Natural Sciences and Engineering Research Council of Canada and Le fond de recherché du Québec, Nature et Technologie.
Ref.: Alexis Vallée-Bélisle, Stephen W Michnick, Visualizing transient protein-folding intermediates by tryptophan-scanning mutagenesis, Nature Structural and Molecular Biology, 2012, DOI: 10.1038/nsmb.2322
