An astounding nanoscale magnified view of bacterial ‘motors’
March 30, 2016
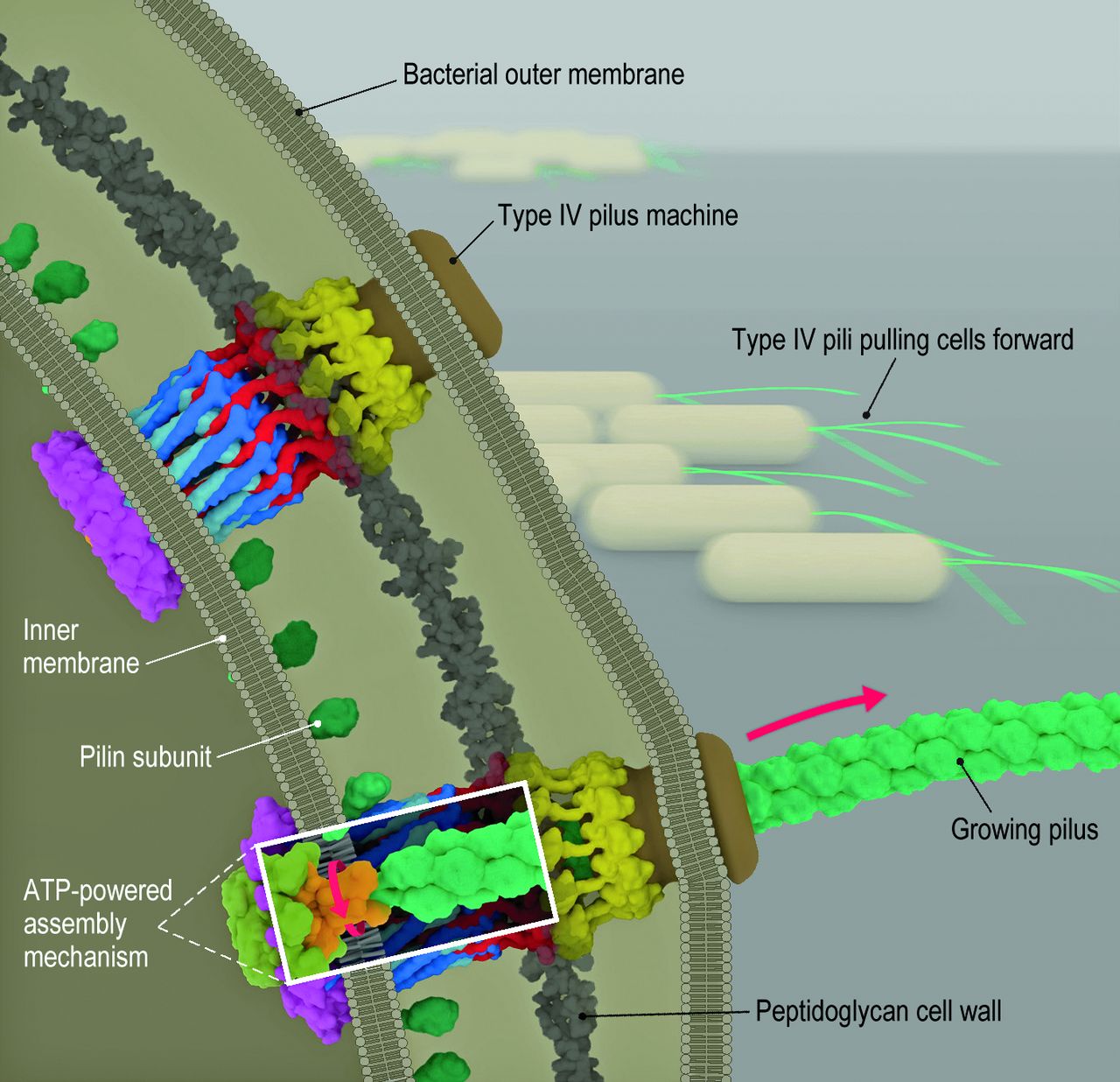
Two bacterial micro machines in a bacterial cell extend and retract fibers (called pili) that pull cells forward. ATP hydrolysis in the cytoplasm generates the energy to cause an assembler-mechanism adapter protein in the inner membrane (detail shown in white rectangle) to rotate, facilitating the transfer of pilin subunits (small green balls) onto the growing pilus. The process is reversed during retraction. (credit: Yi-Wei Chang et al./Science)
Caltech researchers have used a state-of-the-art imaging technique to capture detailed 3D views of the complex mobile nanomachinery in bacteria for the first time.
Grant Jensen, a professor of biophysics and biology at Caltech and an investigator with the Howard Hughes Medical Institute (HHMI), and his colleagues used a technique called electron cryotomography to capture 3D images of intact cells with a resolution that ranges from 2 to 5 nanometers (for comparison, a whole cell can be several thousand nanometers in diameter).

(credit: Sony Pictures/Marvel Studios/HD Wallpapers)
In a paper published in the March 11 issue of the journal Science, the researchers explain they used an advanced technique to analyze bacteria-cell motility machinery — a structure called the type IVa pilus machine (T4PM). This machine enables a bacterium to move through its environment in a manner similar to how Spider-Man travels between skyscrapers.
The T4PM assembles a long fiber (the pilus) that attaches to a surface like a grappling hook and subsequently retracts, thus pulling the cell forward.
Science Magazine | How the bacterium Myxococcus xanthus makes a grappling hook for propulsion
The making of the bacteria mobility movie
First, the researchers froze the bacteria cells instantaneously to prevent water molecules from rearranging to form ice crystals. That locked the cells in place without damaging their structure. Then, using a transmission electron microscope, the researchers imaged the cells from different angles, producing a series of 2-D images. They then digitally reconstructed these images into a 3-D picture of the cell’s structures (like creating a CT scan). Jensen’s laboratory is one of only a few in the entire world that can do this type of imaging.
The researchers found that the bacteria structure is made up of several parts, including a pore on the outer membrane of the cell, four interconnected ring structures, and a stemlike structure. By systematically imaging mutants, each of which lacked one of the 10 T4PM core components, and comparing these mutants with normal M. xanthus cells, they mapped the locations of all 10 T4PM core components, providing insights into pilus assembly, structure, and function.
Strongest motor known in nature — how it works
“The machine lets M. xanthus, a predatory bacterium, move across a field to form a ‘wolf pack’ with other M. xanthus cells, and hunt together for other bacteria on which to prey,” Jensen says. “T4PMs … generate forces as high as 150 pN to pull the cell forward, making T4PMs the strongest molecular motors known,” the authors note.
Another way that bacteria move about their environment is by employing a flagellum — a long whiplike structure that extends outward from the cell. The flagellum is spun by cellular machinery, creating a sort of propeller that motors the bacterium through a substrate (see Tiny swimming ‘biobots’ propelled by heart cells or magnetic fields).
However, cells that must push through the thick mucus of the intestine, for example, need more powerful versions of these motors, compared to cells that only need enough propeller power to travel through a pool of water.
Heavy-duty vs. light-duty bacterial propellers
In a second work, published in an open-access paper in the online early edition of the Proceedings of the National Academy of Sciences (PNAS) on March 14, Jensen and his colleagues used electron cryotomography to study the differences between heavy-duty and light-duty versions of the bacterial propeller. The 3-D images they captured showed that the varying levels of propeller power among several different species of bacteria can be explained by structural differences in these tiny motors.
In order for the flagellum to act as a propeller, structures in the cell’s motor must apply torque — the force needed to cause an object to rotate—to the flagellum. The researchers found that the high-power motors have additional torque-generating protein complexes that are found at a relatively wide radius from the flagellum. This extra distance provides greater leverage to rotate the flagellum, thus generating greater torque. The strength of the cell’s motor was directly correlated with the number of these torque-generating complexes in the cell.
“These two studies establish a technique for solving the complete structures of large macromolecular complexes in situ, or inside intact cells,” Jensen says. “Other structure determination methods, such as X-ray crystallography, require complexes to be purified out of cells, resulting in loss of components and possible contamination. On the other hand, traditional 2-D imaging alone doesn’t let you see where individual protein pieces fit in the complete structure. Our electron cryotomography technique is a good solution because it can be used to look at the whole cell, providing a complete picture of the architecture and location of these structures.”
The work involving the type IVa pilus machinery was published in a Science paper by research scientists at Caltech, the Max Planck Institute for Terrestrial Microbiology, and the University of Utah. The study was funded by the National Institutes of Health (NIH), HHMI, the Max Planck Society, and the Deutsche Forschungsgemeinschaft.
Work involving the flagellum machinery in the second study was published in a PNAS paper. Additional coauthors include collaborators from Imperial College London; the University of Texas Southwestern Medical Center; and the University of Wisconsin–Madison. The study was supported by funding from the UK’s Biotechnology and Biological Sciences Research Council and from HHMI and NIH.
UPDATE April 1, 2016: Added quote from Science paper regarding forces generated by the type IVa pilus machine.
Abstract of Architecture of the type IVa pilus machine
Many bacteria, including important pathogens, move by projecting grappling-hook–like extensions called type IV pili from their cell bodies. After these pili attach to other cells or objects in their environment, the bacteria retract the pili to pull themselves forward. Chang et al. used electron cryotomography of intact cells to image the protein machines that extend and retract the pili, revealing where each protein component resides. Putting the known structures of the individual proteins in place like pieces of a three-dimensional puzzle revealed insights into how the machine works, including evidence that ATP hydrolysis by cytoplasmic motors rotates a membrane-embedded adaptor that slips pilin subunits back and forth from the membrane onto the pilus.
Abstract of Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold
Although it is known that diverse bacterial flagellar motors produce different torques, the mechanism underlying torque variation is unknown. To understand this difference better, we combined genetic analyses with electron cryo-tomography subtomogram averaging to determine in situ structures of flagellar motors that produce different torques, from Campylobacter and Vibrio species. For the first time, to our knowledge, our results unambiguously locate the torque-generating stator complexes and show that diverse high-torque motors use variants of an ancestrally related family of structures to scaffold incorporation of additional stator complexes at wider radii from the axial driveshaft than in the model enteric motor. We identify the protein components of these additional scaffold structures and elucidate their sequential assembly, demonstrating that they are required for stator-complex incorporation. These proteins are widespread, suggesting that different bacteria have tailored torques to specific environments by scaffolding alternative stator placement and number. Our results quantitatively account for different motor torques, complete the assignment of the locations of the major flagellar components, and provide crucial constraints for understanding mechanisms of torque generation and the evolution of multiprotein complexes.
