Editing the genome
July 19, 2011
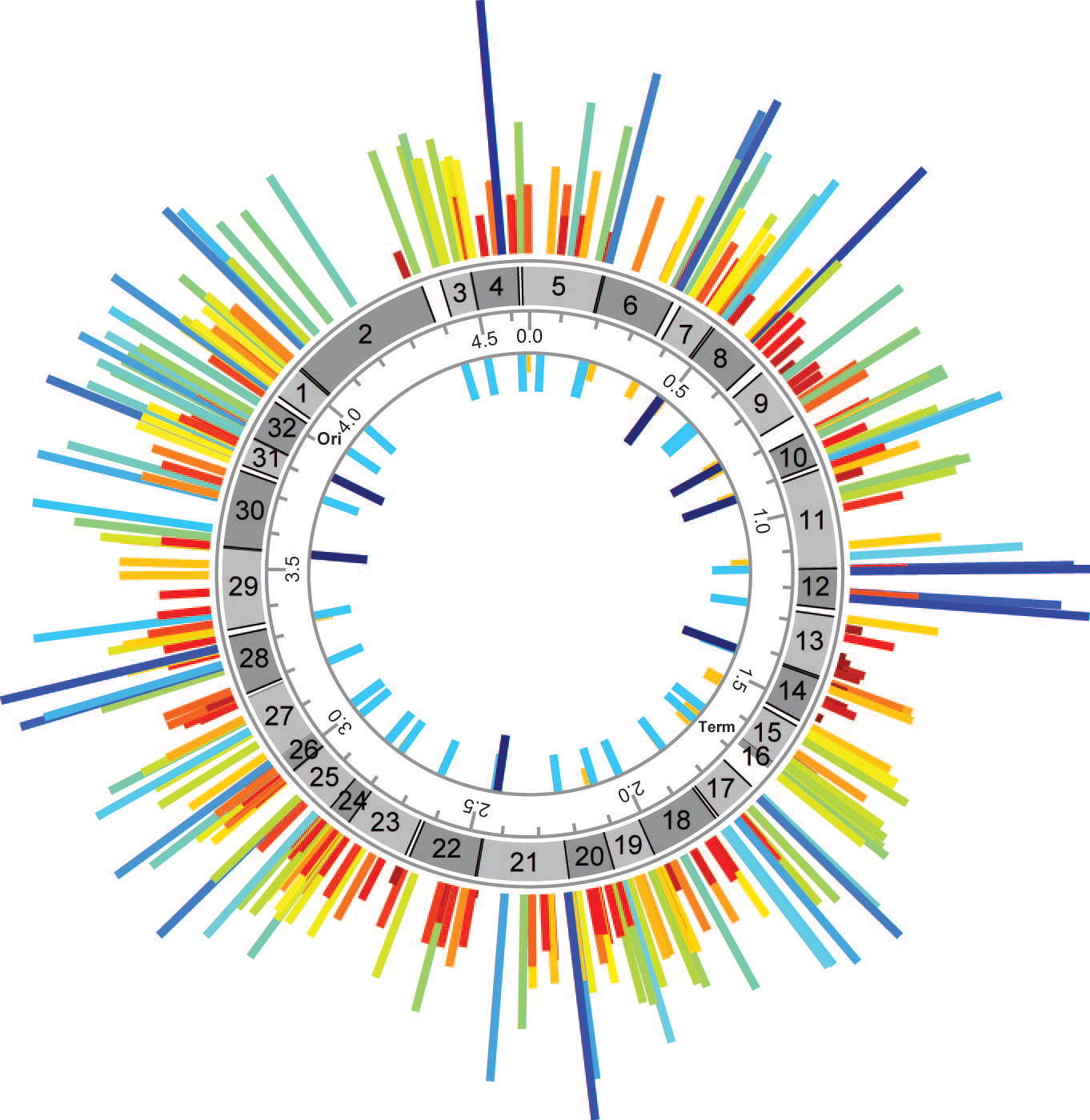
Frequency map of MAGE-generated TAG::TAA codon replacements across the E. coli genome at each TAG codon replacement position. Frequency of TAG::TAA replacements by MAGE across all TAG codons denoted by height- and color-coded bars. (Credit: F.J. Isaacs et. al.)
Researchers at Harvard University Medical School have rewritten the genome of living cells using the genetic equivalent of search and replace — and combined those rewrites in novel cell strains, strikingly different from their forebears.
“The payoff doesn’t really come from making a copy of something that already exists,” said George Church, a professor of genetics at Harvard Medical School who led the research effort in collaboration with Joe Jacobson, an associate professor at the Media Lab at the Massachusetts Institute of Technology. “You have to change it—functionally and radically.”
The researchers are developing genome-scale editing tools that are as fast and easy to use as word processing. For example, they replaced instances of a codon (a DNA word of three nucleotide letters) in 32 strains of E. coli, and then coaxed those partially-edited strains along an evolutionary path toward a single cell line in which all 314 instances of the codon had been replaced.
That many edits surpasses current methods by two orders of magnitude, the researchers said. Using a platform called multiplex automated genome engineering, or MAGE, the team replaced instances of a TAG codon (one that signals “stop”) with another stop codon, TAA, in living E. coli cells.
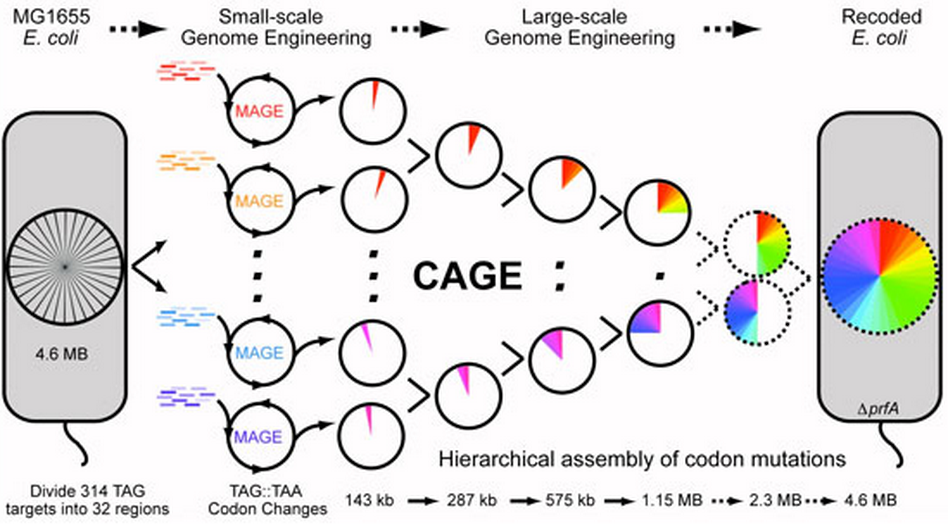
Multiplex automated genome engineering (MAGE) was used to convert all TAG codons to TAA codons. Hierarchical conjugative assembly genome engineering (CAGE) was used to assemble codon changes into higher ordered strains. (Credit: F.J. Isaacs et. al.)
The MAGE process has been called an “evolution machine” for its ability to accelerate targeted genetic change in living cells. The team constructed 32 strains that, taken together, included every possible TAA replacement. Using bacteria’s innate ability to trade genes through conjugation, the researchers induced the cells to transfer genes containing TAA codons at increasingly larger scales. The new method, called conjugative assembly genome engineering, or CAGE, resembles a playoff bracket — a hierarchy that winnows 16 pairs to eight to four to two to one.
Their results suggested that the final four strains were healthy (in spite of skepticism from colleagues), as the team assembled four groups of 80 engineered alterations into stretches of the chromosome surpassing one million DNA base pairs.
Ref.: Farren J. Isaac, et al., Precise Manipulation of Chromosomes in Vivo Enables Genome-Wide Codon Replacement, Science, 2011; Vol. 333 no. 6040 pp. 348-353, [DOI: 10.1126/science.1205822]
