How to create the world’s most complex 3D-motion nanomachines from DNA
January 12, 2015
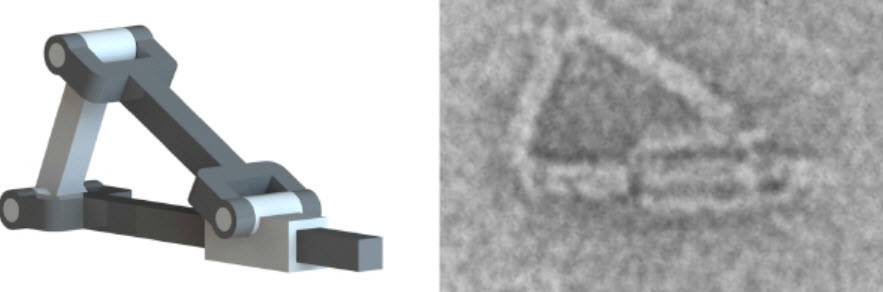
A machine design (left) and a nanoscale emulation using DNA origami (transmission electron microscope image: right) (credit: The Ohio State University)
Mechanical engineers at The Ohio State University have designed and constructed complex nanoscale mechanical parts using “DNA origami” — proving that the same basic design principles that apply to typical full-size machine parts can now also be applied to DNA — and can produce complex, controllable components for future nanorobots.
In a paper in Proceedings of the National Academy of Sciences, the engineers describe how they used a combination of natural and synthetic DNA to build complex machines that can perform repetitive tasks.
The DNA origami design principle is also seen in this character from “Transformers: Revenge of the Fallen” (credit: Paramount Pictures)
“Nature has produced incredibly complex molecular machines at the nanoscale, and a major goal of bio-nanotechnology is to reproduce their function synthetically,” said project leader Carlos Castro, assistant professor of mechanical and aerospace engineering. “Where most research groups approach this problem from a biomimetic standpoint — mimicking the structure of a biological system — we decided to tap into the well-established field of macroscopic machine design for inspiration.”
“In essence, we are using a bio-molecular system to mimic large-scale engineering systems to achieve the same goal of developing molecular machines,” he said.
Ultimately, the technology could create complex nanorobots to deliver medicine inside the body or perform nanoscale biological measurements, among many other applications. Like the fictional “Transformers,” DNA origami machine could change shape for different tasks.
“I do think we can ultimately build something like a Transformer system, though maybe not quite like in the movies. I think of it more as a nano-machine that can detect signals such as the binding of a biomolecule, process information based on those signals, and then respond accordingly—maybe by generating a force or changing shape.”
How to build a complex nanomachine
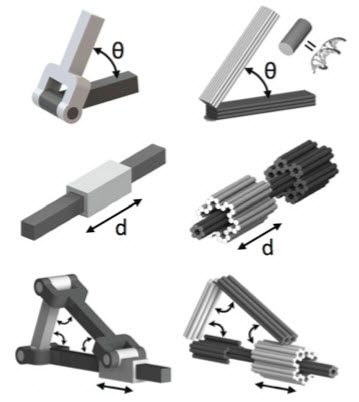
DNA origami mechanism design approach, with rotational and linear motion. Left: macroscale solid models. Right: DNA origami counterpart models (cylinders represent double-strand DNA helices). (Credit: Alexander E. Marras/PNAS)
The DNA origami method for making nano-structures has been widely used since 2006, and is now a standard procedure for many labs that are developing future drug delivery systems and electronics. It involves taking long strands of DNA and coaxing them to fold into different shapes, then securing certain parts together with “staples” made from shorter DNA strands.
The resulting structure is stable enough to perform a basic task, such as carrying a small amount of medicine inside a container-like DNA structure and opening the container to release it.
To create more complex nanomachines that could perform such tasks repeatedly, Castro joined with Haijun Su, also an assistant professor of mechanical and aerospace engineering at Ohio State. Combined, the two research teams have expertise in nanotechnology, biomechanics, machine engineering and robotics.
Two basic methods
There are two keys to their unique approach for designing and controlling the machines’ motion, Castro explains:
- Make certain parts of the structure flexible using single-stranded DNA and stiffer using double-stranded DNA.
- “Tune” the DNA structures so that the machines’ movements are reversible and repeatable. For that, the researchers dot their structures with synthetic DNA strands that hang off the edges. These strands are designed to act like strips of hook and loop fasteners — they stick together or unstick depending on chemical cues from the machine’s surroundings.
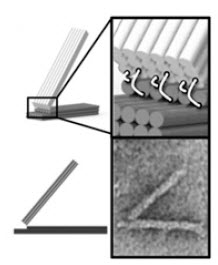
DNA origami hinge, consisting of two stiff bundles of 18 double-strand DNA helices connected at one end by six flexible single-strand DNA connectors (white wires) (TEM image shown below) (credit: Alexander E. Marras/PNAS)
In the lab, doctoral students Alexander Marras and Lifeng Zhou took long strands of DNA from a bacteriophage — a virus that infects bacteria and is harmless to humans — and “stapled” the strands to short strands of synthetic DNA. First, they joined two stiff DNA “planks” with flexible staples along one edge to create a simple hinge. Castro likened the process to “connecting two wooden 2×4’s with very short pieces of string along the 4-inch edge at one end.”
A 3D motion machine
They also built a system that moved a piston inside a cylinder. That machine used five planks, three hinges and two tubes of different diameters—all made from pieces of double-stranded and single-stranded DNA.
To test whether the machines were moving properly, they imaged them with transmission electron microscopy (TEM). They also labeled the DNA with fluorescent tags, so that they could observe the shape changes with a spectrofluorometer.
Tests confirmed that the hinges opened and closed and the piston moved back and forth, and that researchers could control the motion with the addition of chemical cues to the solution, such as additional strands of DNA.
This approach of designing simple joints and connecting them together to make more complex working systems is common in macroscopic machine design, but the engineers say this is the first time it’s been done with DNA, and the first time anyone has tuned the DNA to produce reversible actuation of a complex mechanism.
This research was funded by the National Science Foundation (NSF) and the Center for Emergent Materials at Ohio State. The Center for Emergent Materials is an NSF Materials Research Science and Engineering Center. Microscopy access was provided by the Campus Microscopy and Imaging Facility and the Center for Electron Microscopy and Analysis at Ohio State, as well as the Department of Physics.
The Ohio State University | A DNA “origami” machine created at The Ohio State University.
The Ohio State University | A hinge created with DNA “origami” at The Ohio State University.
Abstract of Programmable motion of DNA origami mechanisms
DNA origami enables the precise fabrication of nanoscale geometries. We demonstrate an approach to engineer complex and reversible motion of nanoscale DNA origami machine elements. We first design, fabricate, and characterize the mechanical behavior of flexible DNA origami rotational and linear joints that integrate stiff double-stranded DNA components and flexible single-stranded DNA components to constrain motion along a single degree of freedom and demonstrate the ability to tune the flexibility and range of motion. Multiple joints with simple 1D motion were then integrated into higher order mechanisms. One mechanism is a crank–slider that couples rotational and linear motion, and the other is a Bennett linkage that moves between a compacted bundle and an expanded frame configuration with a constrained 3D motion path. Finally, we demonstrate distributed actuation of the linkage using DNA input strands to achieve reversible conformational changes of the entire structure on ∼minute timescales. Our results demonstrate programmable motion of 2D and 3D DNA origami mechanisms constructed following a macroscopic machine design approach.
